/
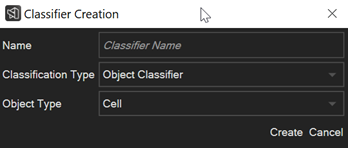
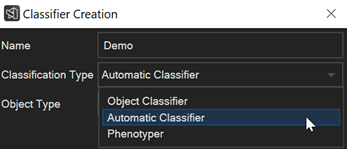
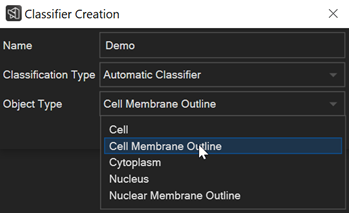
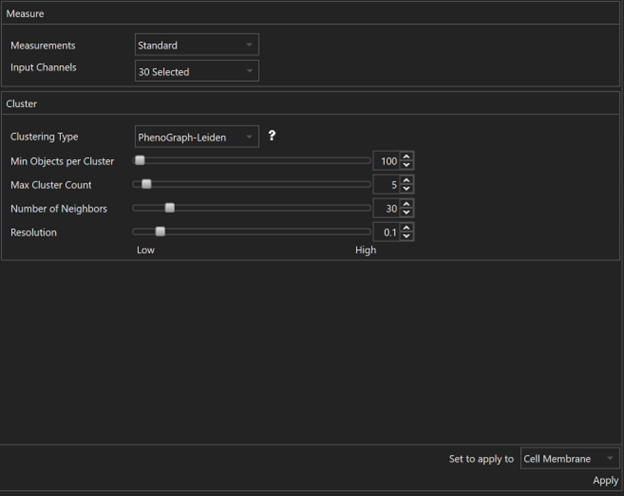
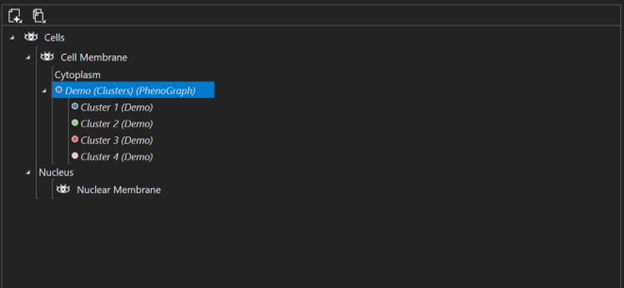
How to auto-classify objects and visualize them
Aivia Software
How to auto-classify objects and visualize them
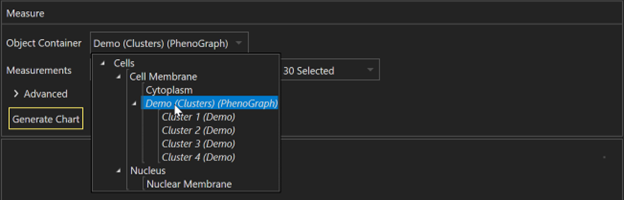
, multiple selections available,
Related content
3D Multiplexed Cell Detection Recipe
3D Multiplexed Cell Detection Recipe
More like this
Phenotyper
Phenotyper
More like this
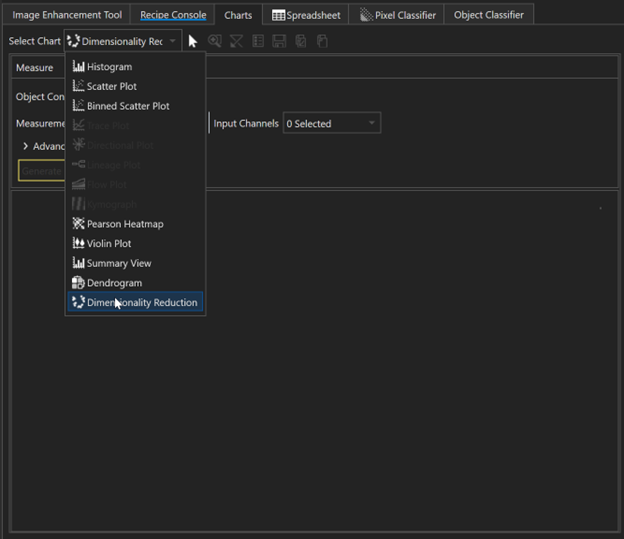
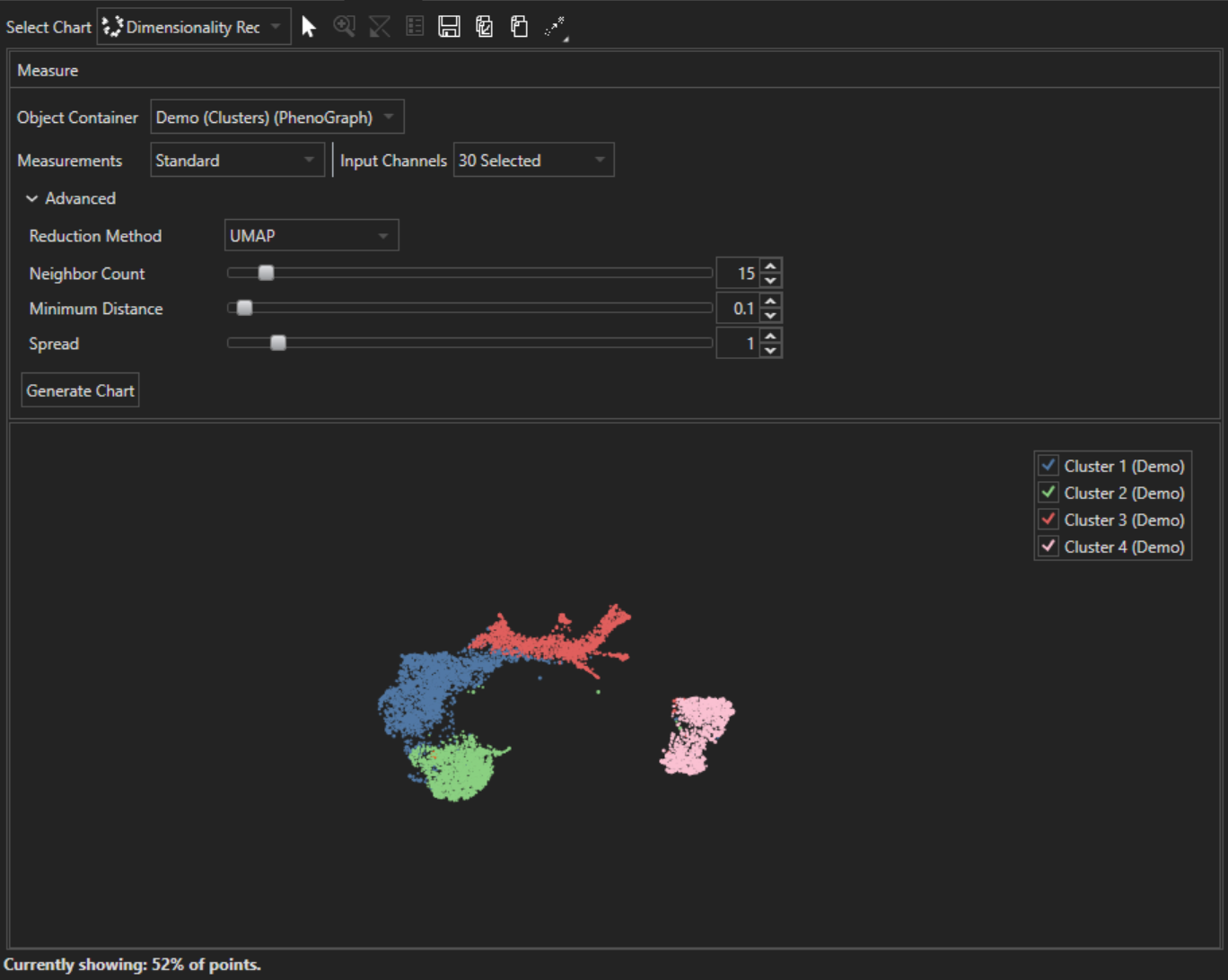
Dimensionality Reduction
Dimensionality Reduction
Read with this
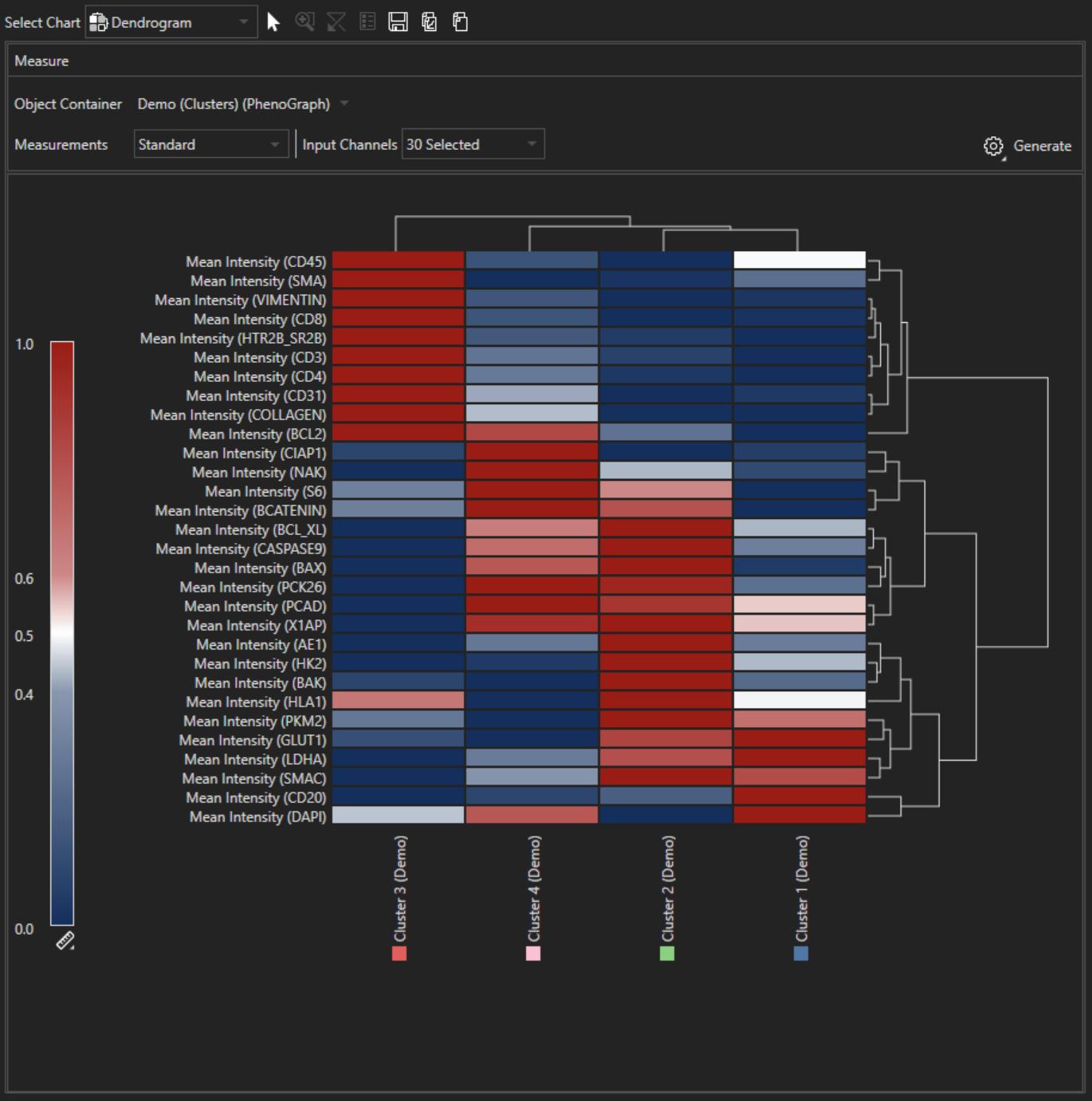
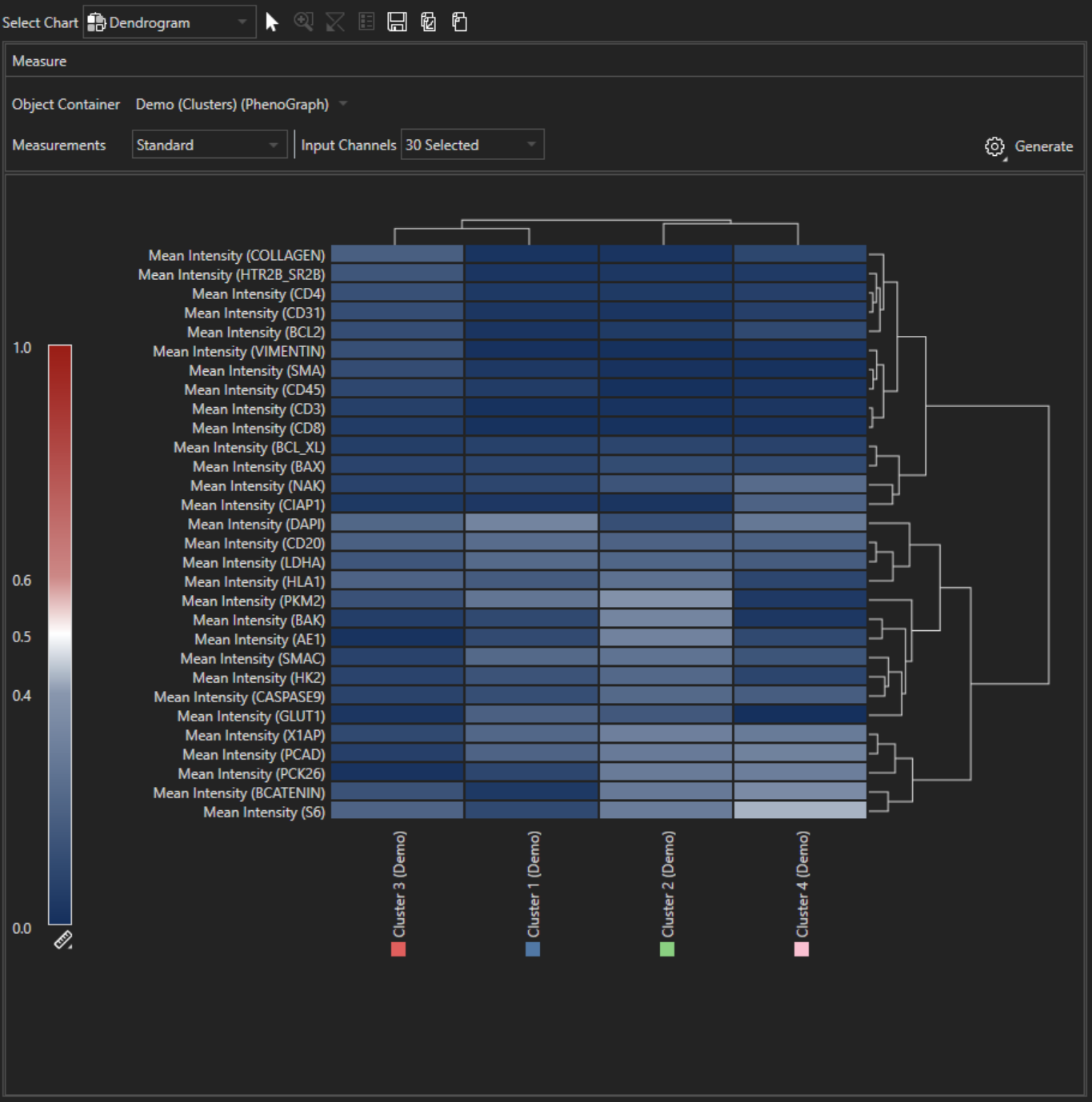
Dendrogram
Dendrogram
Read with this
Automatic Object Classifier
Automatic Object Classifier
More like this
User-guided Classifier
User-guided Classifier
More like this