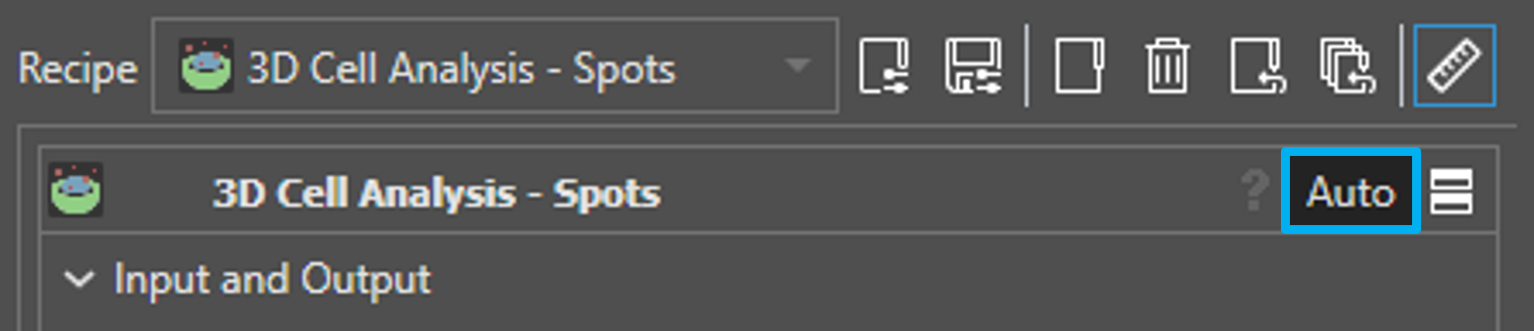
Automatic Parameter Selection
3D Cell Analysis - Meshes/Spots features automatic parameter selection for Min Cells Intensity, Min Nuclei Intensity, and Min Vesicles Intensity. Click on Auto (see figure below) in the recipe header to initiate parameter estimation, which uses the image data in the processing region as well as the smoothing and background removal options that you have selected (see “Parameters” section) to approximate reasonable minimum intensity values.
Parameters and Presets
Parameters
Recipe parameters for 3D Cell Analysis - Meshes and 3D Cell Analysis - Spots and their descriptions are summarized in the table below.
Preset Group | Parameter Name | Min Value | Max Value | Description |
|---|---|---|---|---|
Whole Cells Detection | Image Smoothing Filter Size (without Skip Smooth Image only) | 1 | 100 | Specifies the diameter of the filter that is used to smooth the Cells Input before further processing The available smoothing types are the following:
|
Typical Cells Diameter (Remove Background only) | 0 | 65,535 ( px or µm) | Specifies the diameter of a typical cell in the image; the diameter is used for object enhancement and background removal, with a lower value preserving smaller objects | |
Min Cells Intensity | 0 | 255 (8-bit) 65,535 (16-bit) | Specifies the lower intensity threshold for cell detection
A lower value leads to the detection of bigger and more cells | |
Fill Holes Size | 0 | 1,000,000 ( px2 or µm2) | Specifies the maximum size of gaps in detected cells that are filled; a lower value leads to the preservation of more holes in the detected cells | |
Cell Diameter | 0 | 100,000 ( px or µm) | Specifies the range of detected cells to keep based on their diameters | |
Min Edge to Center Distance (Apply Partition only) | 0 | 100,000 ( px or µm) | Specifies the minimum distance from the center of a cell to the edge that is touching its closest neighboring cell; a lower value leads to more aggressive partitioning, which results in smaller, more uniform cell objects | |
Mesh Smoothing Factor | 0 | 100 | Specifies the amount of smoothing applied to the surface reconstructions of the detected cells; a lower value leads to the creation of surfaces with greater similarity to the input image | |
Nuclei Detection (optional) | Image Smoothing Filter Size (without Skip Smooth Image only) | 1 | 100 | Specifies the diameter of the filter that is used to smooth the Nuclei Input before further processing The available smoothing types are the following:
|
Typical Nuclei Diameter (Remove Background only) | 0 | 100,000 ( px or µm) | Specifies the diameter of a typical nucleus in the image; the diameter is used for object enhancement and background removal, with a lower value preserving smaller objects | |
Min Nuclei Intensity | 0 | 255 (8-bit) 65,535 (16-bit) | Specifies the lower intensity threshold for nuclei detection
A lower value leads to the detection of bigger and more nuclei | |
Fill Holes Size | 0 | 1,000,000 ( px2 or µm2) | Specifies the maximum size of gaps in detected nuclei that are filled; a lower value leads to the preservation of more holes in the detected nuclei | |
Nuclei Diameter | 0 | 100,000 ( px or µm) | Specifies the range of detected nuclei to keep based on their diameters | |
Min Edge to Center Distance (Apply Partition only) | 0 | 100,000 ( px or µm) | Specifies the minimum distance from the center of a nucleus to the edge that is touching its closest neighboring nucleus; a lower value leads to more aggressive partitioning, which results in smaller, more uniform nucleus objects | |
Mesh Smoothing Factor | 0 | 100 | Specifies the amount of smoothing applied to the surface reconstructions of the detected nuclei; a lower value leads to the creation of surfaces with greater similarity to the input image | |
Nuclei Correction | Not applicable | Specifies how to deal with detected nuclei that lie partially outside of detected cells
| ||
Vesicles [#] Detection Up to five (5) vesicle sets can be detected. (optional) | Image Smoothing Filter Size (without Skip Smooth Image only) | 1 | 100 | Specifies the diameter of the filter that is used to smooth the Vesicles [#] Input before further processing The available smoothing types are the following:
|
Typical Vesicles Diameter (Remove Background only) | 0 | 100,000 ( px or µm) | Specifies the diameter of a typical vesicle in the image; the diameter is used for object enhancement and background removal, with a lower value preserving smaller objects | |
Min Vesicles Intensity | 0 | 255 (8-bit) 65,535 (16-bit) | Specifies the lower intensity threshold for vesicle detection
A lower value leads to the detection of bigger and more vesicles | |
Fill Holes Size | 0 | 1,000,000 ( px2 or µm2) | Specifies the maximum size of gaps in detected vesicles that are filled; a lower value leads to the preservation of more holes in the detected vesicles | |
Vesicles Diameter | 0 | 100,000 ( px or µm) | Specifies the range of detected vesicles to keep based on their diameters | |
Min Edge to Center Distance (Apply Partition only) | 0 | 100,000 ( px or µm) | Specifies the minimum distance from the center of a vesicle to the edge that is touching its closest neighboring vesicle; a lower value leads to more aggressive partitioning, which results in smaller, more uniform vesicle objects | |
Mesh Smoothing Factor (3D Cell Analysis - Meshes only) | 0 | 100 | Specifies the amount of smoothing applied to the surface reconstructions of the detected vesicles; a lower value leads to the creation of surfaces with greater similarity to the input image | |
Measurements
The 3D Cell Analysis - Meshes and 3D Cell Analysis - Spots recipes generate a few morphological, intensity, position, and count measurements. You can add additional measurements to the analysis results by using the Measurement Tool in Aivia and explore measurement definitions on the Measurement Definitions page. The measurements generated by the recipe are in the table below.
Object Set | Morphology | Intensity | Position | Count |
|---|---|---|---|---|
Cell Membranes |
|
|
|
|
Nuclear Membranes |
|
|
|
|
Cells |
| None |
|
|
Cytoplasm |
| None |
|
|
Nucleus |
| None |
|
|
Vesicles |
| None |
| None |